Genetics and neurobiology of C. elegans
Principal investigator: Jean-Louis BESSEREAU
C. elegans | synapse | neuromuscular junction | GABA receptors | Acetylcholine receptors | aging | genetics | molecular neurobiology | electrophysiology | optogenetics | super-resolution microscopy | electron microscopy
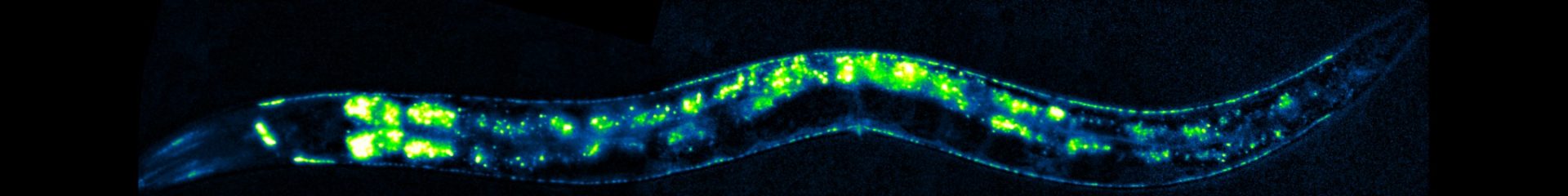
Synapses are sophisticated nanomachines that support transfer and processing of information between excitable cells.
Since most neurons receive thousands of synaptic inputs, the neuronal membrane is a mosaic of specialized microdomains where neurotransmitter receptors cluster in register with the corresponding presynaptic neurotransmitter release sites. Our lab is interested in identifying the cellular and molecular mechanisms involved in the organization and maintenance of the synapse with a specific focus on the control of neurotransmitter receptor expression and localization.
Our strategy is based on the combination of genetics, imaging, electrophysiology and biochemistry in the nematode Caenorhabditis elegans (for more information on C. elegans see “an overview of the model organism C. elegans“). Using the neuromuscular synapse as a model synapse, we identified several new genes involved in the clustering of acetylcholine and GABAA receptors through previously undescribed mechanisms, including a novel anterograde synaptic organizer that assembles extracellular scaffolds in the synaptic cleft. We are currently analyzing the organization, dynamics and maintenance of these synaptic scaffolds as well as the genes involved in the biosynthesis and trafficking of the receptors.
Our results should contribute to a better understanding of the normal and pathological synapse. Specifically, synaptic defects have been involved over the last years in the pathogenesis of a growing number of neuropsychiatric diseases, leading to the concept of “synaptopathies”. However, a number of genes linked to neuropsychiatric diseases have no assigned function, and it is likely that the mutational landscape of these diseases will be complexified by the wealth of data generated with next generation sequencing techniques. Simple organisms should help!
RESTOICH - Ionotropic receptor stoichiometry: pathways and pathology
2022-07-28

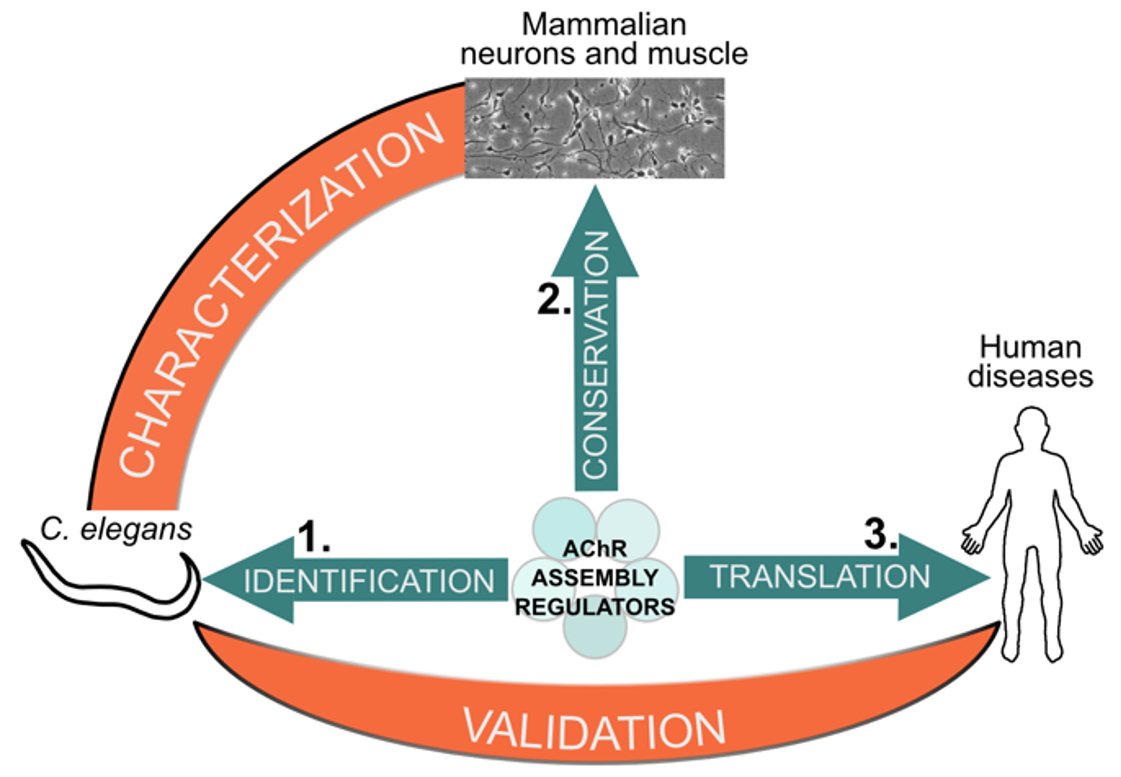
The project of our researcher Manuela D’Alessandro has been selected by ANR JCJC. We will investigate the cellular principles that govern the stoichiometric composition of AChRs, in particular through the identification of novel factors that control AChR composition and their implication in neuromuscular diseases.
The large number of neuronal AChRs subtypes, together with heterogeneity in cellular and subcellular localization, hinder the research on mechanisms controlling receptor stoichiometry using traditional biochemical strategies.
Because AChRs have been extremely conserved during evolution, we will address this question first using the nematode C. elegans as a genetically tractable system, and then in human cells. We will also use the C. elegans model to rapidly validate novel variants in genes controlling AChR assembly from undiagnosed patients and assess their severity.
Genetic control of acetylcholine receptor expression: from new mechanisms to functional genomics
2022-08-31

Our project on genetic control of AChR expression has been selected by AFM. We will try to get a comprehensive genetic landscape of We will perform new-generation screens for decreased amount of AChR at the
C. elegans neuromuscular junction based on direct visualization of the receptors in vivo. Then we will test the functional conservation of
C. elegans genes for AChR biosynthesis in mammals using advanced genome manipulation in mammalian cell lines. Finally we have developed a pipeline to go back from human gene variants to C. elegans and we will assess the pathogenicity of human polymorphisms found in genes required for AChR biosynthesis.
modulators and shed new light on the cell biology of AChR.











