Post-doctorant – équipe ACDC .
Context
Cerebral glial cells, particularly microglia and astrocytes, play a key role in neurological and psychiatric disorders through neuroinflammation. While their activation marks disease progression, it remains unclear whether inhibiting inflammation helps or harms patients. Current imaging of glial activation relies on PET targeting the 18-kDa translocator protein, but this method is costly, limited in availability, and involves radiation. There is an urgent need for non-invasive, sensitive imaging alternatives. The white matter, due to its organized structure and low vascular density, may offer a better environment than gray matter for developing a diffusion MRI (dMRI)-based method to detect cerebral inflammation.
Related publication: Benkeder et al, Elife, 2025. DOI: 10.7554/eLife.101630
Aims of the project
The global aim of this collaborative project financed by ANR is to develop a specific dMRI-based method for detecting cerebral inflammation using a machine learning classifier. This classifier will be trained with Monte-Carlo simulations of dMRI signals obtained from digital phantoms of the complex white matter microstructure, including axons, resting and activated microglia, and astrocytes. The purpose of this position is to characterize the morphology of microglia and astrocyte in various neuroinflammatory conditions using light microscopy.
Main tasks
Characterize the morphology of microglia and astrocytes in various neuroinflammatory conditions, such as neuromyelitis optica and multiple sclerosis, using animal models. Perform brain tissue sectioning, immunohistochemistry, and confocal imaging to acquire high-resolution morphological data on glial cells. Develop automated image analysis pipelines using Python or MATLAB to quantify the morphological features of microglia and astrocytes. Provide the obtained morphological metrics to the project collaborators to be incorporated into the computational model for dMRI-based neuroinflammation detection.
Qualifications
PhD candidate or engineer in bioinformatics, computational biology, neuroscience, or a related field. Proficient in programming and data analysis, with experience in Python or MATLAB. Familiarity with image processing and analysis techniques, such as segmentation and feature extraction. Excellent communication skills in English (B2-C1 level) for scientific writing and presentations.
Conditions
3-year contract.
Location: Lyon, France. MeLiS laboratory link
Stimulating research environment with access to state-of-the-art technological platforms.Application
Candidates should email a CV, cover letter and contact details of two references to olivier.pascual@inserm.fr. Applications will be accepted until 20/12/2025.
Project
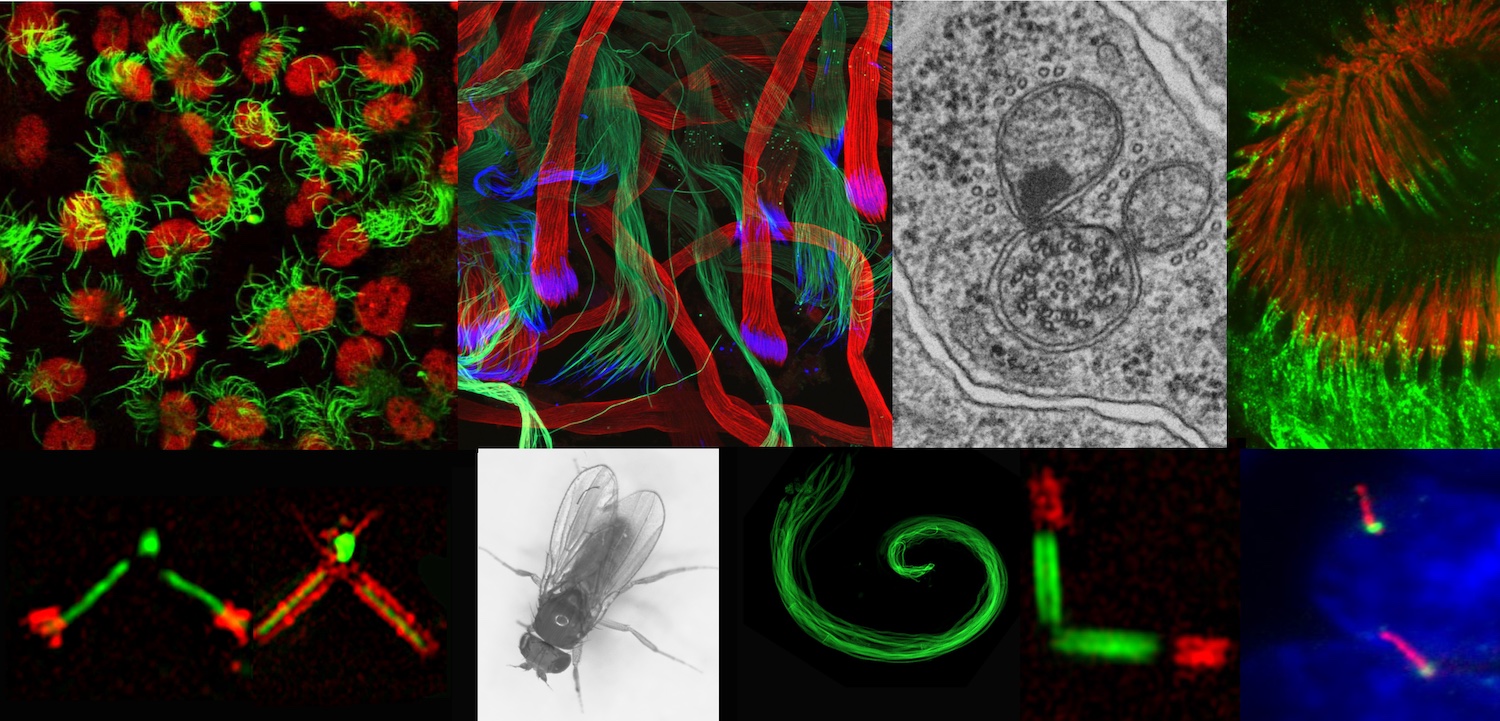
The candidate (M/F) will be recruited as part of an ANR-funded project conducted in collaboration with a laboratory at the University of Strasbourg. The project aims to understand the molecular mechanisms of ciliogenesis in general and in the specific context of photoreceptors.
Cilia are tiny microtubular organelles found on the surface of most mammalian cells. Alterations in their formation, stability or function cause rare diseases known as ciliopathies. These are associated with a wide range of symptoms, many of which involve abnormalities in photoreceptors (PR) leading to retinal degeneration. PR homeostasis depends on the correct assembly and function of their cilia, but many different mechanisms have been implicated in the retinal degeneration associated with ciliopathies.
Aims of the project
The objectives of the project are 1) to identify new players in ciliary homeostasis using biochemical and proteomic approaches on mammalian cells, and 2) to study the molecular mechanisms of ciliogenesis involving the proteins identified both in cultured cells and in the context of PRs by combining functional tests and microscopy-based approaches, in particular high-resolution expansion microscopy (U-ExM).
Main tasks
- Using biochemical and proteomic approaches, identify proteins associated with centrioles/cilia (generate the centriole proteome by Capture (10.1016/j.devcel.2023.09.008) or sucrose gradient approaches; immunoprecipitation; molecular dissection of protein-protein interactions),
- Apply molecular biology, cell biology and imaging approaches to study the roles of the identified proteins in ciliogenesis (cell culture, mouse retina).
Qualifications
PhD in biochemistry or equivalent. Experience in biochemistry and proteomics is required. Experience in microscopy is a plus. If not, willingness to acquire expertise in microscopic approaches is mandatory. Fluent in both spoken and written English and with good communication skills.
Working environment
The project will be carried out within the MeLiS unit in Prof. Bénédicte Durand's team under the supervision of Dr. Véronique Morel, ANR project leader. The team studies the molecular processes of ciliogenesis, using Drosophila and cultured cells as model systems and developing approaches in genetics, molecular biology and microscopy. The projects draw on extensive expertise in expansion microscopy, an innovative tissue preparation technique that enables super-resolution observations with conventional microscopes, both in cells and thick tissues.
Conditions
1-year contract (renewable)
Location: Lyon, France. MeLiS laboratoryApplication
Candidates should email a CV, cover letter and contact details of two references to veronique.morel@univ-lyon1.fr. Applications will be accepted until 20/12/2025.
Context
Learning and memory involve strengthening specific neuronal assemblies through Hebbian plasticity, allowing their reactivation during recall. Sleep is crucial for memory consolidation, with hippocampal sharp-wave ripples and thalamocortical spindles replaying and transferring information to the cortex for long-term storage. Spindles alone can also reinforce cortical memory traces. While these processes are well-studied at the level of neurons and dendrites, their synaptic underpinnings remain unclear, and the role of microglial cells is still largely unknown.
Related publication: Hristovska et al, Nat Comm, 2022. DOI: 10.1038/s41467-022-34035-9
Aims of the project
To study the impact of mutations on the biosynthesis, assembly and trafficking of potassium channels in vivo using genetically modified models in C. elegans.
This original project combines complementary approaches for an in-depth assessment of the molecular and cellular mechanisms associated with pathogenic variants.Main tasks
Surgery (viral injections, electrode implantation), live two-photon imaging on awake mice, electrophysiology, data treatment, basic biochemistry (WB, PCR, immunohistochemistry).
Qualifications
PPhD in Neuroscience or equivalent with less than two year of experience. Python or Matlab coding is required, FELASA grade B or C is mandatory. Fluent in both spoken and written English (B2-C1) and with good/excellent communication skills.
Conditions
1-year contract (renewable).
Location: Lyon, France. MeLiS laboratory link
Stimulating research environment with access to state-of-the-art technological platforms.Application
Candidates should email a CV, cover letter and contact details of two references to olivier.pascual@inserm.fr. Applications will be accepted until 15/09/2025.
